Artificial intelligence predicts immune and inflammatory genetic signatures directly from hepatocellular carcinoma histology. A study led by Prof. Julien Calderaro and published in Journal of Hepatology
 Immune gene signatures reflecting the quality and quantity of the intra-tumor immune infiltration are known to predict sensitivity to immunotherapy, but their use in the clinical setting is however challenging, as they require access to molecular biology platforms for the extraction of nucleic acids and their processing/sequencing. They are also highly dependent on sample quality and are prone to standardization and bioinformatics expertise issues.
Immune gene signatures reflecting the quality and quantity of the intra-tumor immune infiltration are known to predict sensitivity to immunotherapy, but their use in the clinical setting is however challenging, as they require access to molecular biology platforms for the extraction of nucleic acids and their processing/sequencing. They are also highly dependent on sample quality and are prone to standardization and bioinformatics expertise issues.
On the other hand, histological slides are readily available in pathology departments. It is well established that they contain a large amount of information that allows definitive diagnosis and prediction of clinical outcomes. The advent of digital pathology and artificial intelligence (AI) also allows to standardize their analysis and to extract significant morphological characteristics that are not easily accessible to the human eye.
In a proof-of-concept study led by Pr Julien Calderaro (IMRB_U955, team Jean-Michel Pawlotsky), the authors developed and validated artificial intelligence models capable of phenotyping, from simple digital histological slides, the immune characteristics of liver cancers.
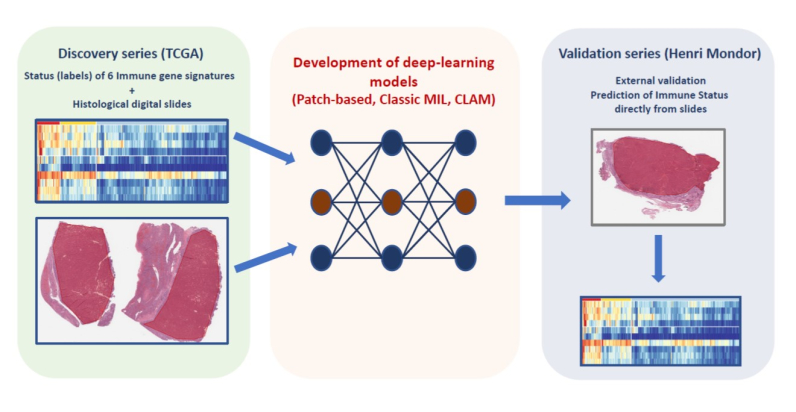
This type of approach could constitute a novel type of biomarker, and could allow a more rapid identification patients who may benefit from immunomodulatory therapies.
This work has been published in Journal of Hepatology (IF 25) February 7, 2022 (full text via Insermbiblio)Artificial intelligence predicts immune and inflammatory gene signatures directly from hepatocellular carcinoma histology.(full text via Insermbiblio)
Zeng Q, Klein C, Caruso S, Maille P, Laleh NG, Sommacale D, Laurent A, Amaddeo G, Gentien D, Rapinat A, Regnault H, Charpy C, Nguyen CT, Tournigand C, Brustia R, Pawlotsky JM, Kather JN, Maiuri MC, Loménie N, Calderaro J.

